Prostate Cancer Detection using GP
Images of healthy tissue (upper panel) and cancerous tissue (lower panel).
This practice is based on the use of Gaussian processes for the classification of histological images of the prostate, according to the existence or not of cancer.
Specifically, this project uses a data set of 1014 images from healthy tissue and 298 from carcinogenic tissue, from which 10 descriptors have been extracted. In this set of data, the folds to be used in cross validation are predetermined, so that the data are first divided according to their class and then each of them in 5 sets of similar size, each representative of a fold of cross validation.
To carry out the classification of the image data, the Gaussian process algorithm will be used, whose functioning will be defined in this report, prior to the presentation of the results obtained through it in the data set. Thus, one of the main challenges of the problem presented here, is the need to achieve the best possible classification through this algorithm, on a set of data that is highly unbalanced. For this reason and for each of the folds of the majority class (negative class) of the cross validation to be performed, we will take all the data of the positive class in order to balance the observations of each class in the models to be trained.
What is a Gaussian process?
A Gaussian process consists of a (potentially infinite) collection of random variables, each of which follows a normal distribution, for which given any finite subset of the same, this follows a multivariate normal distribution.
\[f(x) ∼ GP(m(x), k(x,x'))\]This notation means that for each value \(x ∈ R^n\), we consider the function \(f(x)\) as a random variable, that is to say, a distribution is defined on the set of real functions, being the function of average:
\[m(x) := E[f(x)]\]and the covariance function:
\[k(x, x') := E[(f(x) − m(x))(f(x') − m(x'))]\]Intuitively this means that the value of \(f(x)\) is treated as a random variable with an average of \(m(x)\) and a covariance between pairs of random variables.
Thus, a Gaussian process defines a previous distribution, which can be converted to a later distribution once we have seen some data. Although it may seem difficult to represent a distribution over a function, it turns out that we just need to be able to define it over the function values in a set of finite points, but arbitrary, for example \(x_{1},...,x_{N}\). A Gaussian process assumes that \(p(f(x_{1}),...,f(x_{N}))\) jointly follows a Gaussian, with average \(μ(x)\) and covariance \(∑(x)\) given by \(∑_{i,j}=k(x_{i},x_{j})\), where $k$ is a kernel function. The key idea is that if \(x_{i}\) and \(x_{j}\) the kernel considers them to be similar, then we expect the output of the function at those points to be similar as well.
Just as for \(μ(x)\) we can use any real function, for \(k(x_{i},x_{j})\) it must be fulfilled that for any set of elements, the resulting matrix is valid for a multivariable Gaussian distribution (semi-definitive positive), which are the same conditions as for kernels, so any kernel function can be used as a covariance function.
Software used
In order to carry out the present project, the programming language Python has been used, in its version 3.7. Similarly, the development environment used is Pycharm, from the software company JetBrains. Among the different Python packages in which the Gaussian process algorithm is implemented, in our case we will use GPflow. GPflow is a package for the construction of Gaussian process models in python, using TensorFlow to execute the calculations, which allows a faster execution than the rest of packages, because it can make use of GPU.
To implement the models used in this project, the object VGP, present in the models package, has been used. This implementation of the Variational Gaussian Process (VGP) approximates the subsequent distribution to a multivariate Gaussian. On these VGP models, we have defined the probability distribution that we will use. In our case, as it is a binary classification problem, we will use a Bernouilli distribution. Finally, according to the formal requirements established for this work, two different types of kernel are used for the Gaussian process: one linear (Linear) and another Gaussian (RBF), both within the kernels package. For both kernels the size of the input_dim variable has been modified to set it to the number of variables to consider of our data set.
In addition, the other software packages that have been used for the implementation of this practice have been:
- matplotlib: To carry out the visualizations of the graphics to be shown in the results (ROC curves, confusion matrices…).
- Sklearn: Both to carry out pre-processing tasks on the data and to calculate metrics to evaluate the classification made by the generated models.
- numpy: To carry out calculations on matrices in a simpler and more efficient way.
Experimental results
The results obtained for the cross validation of Gaussian process models are shown below, first with the linear kernel and then with the Gaussian.
Linear Kernel
After cross validation on the data set, using Gaussian processes with linear kernel, we have obtained a series of metrics, which are presented below.
ROC curve and precision-sensitivity curve
It presents both the ROC curves obtained for each of the models generated in the different divisions of the data in the cross validation, as well as the precision and sensitivity curves.
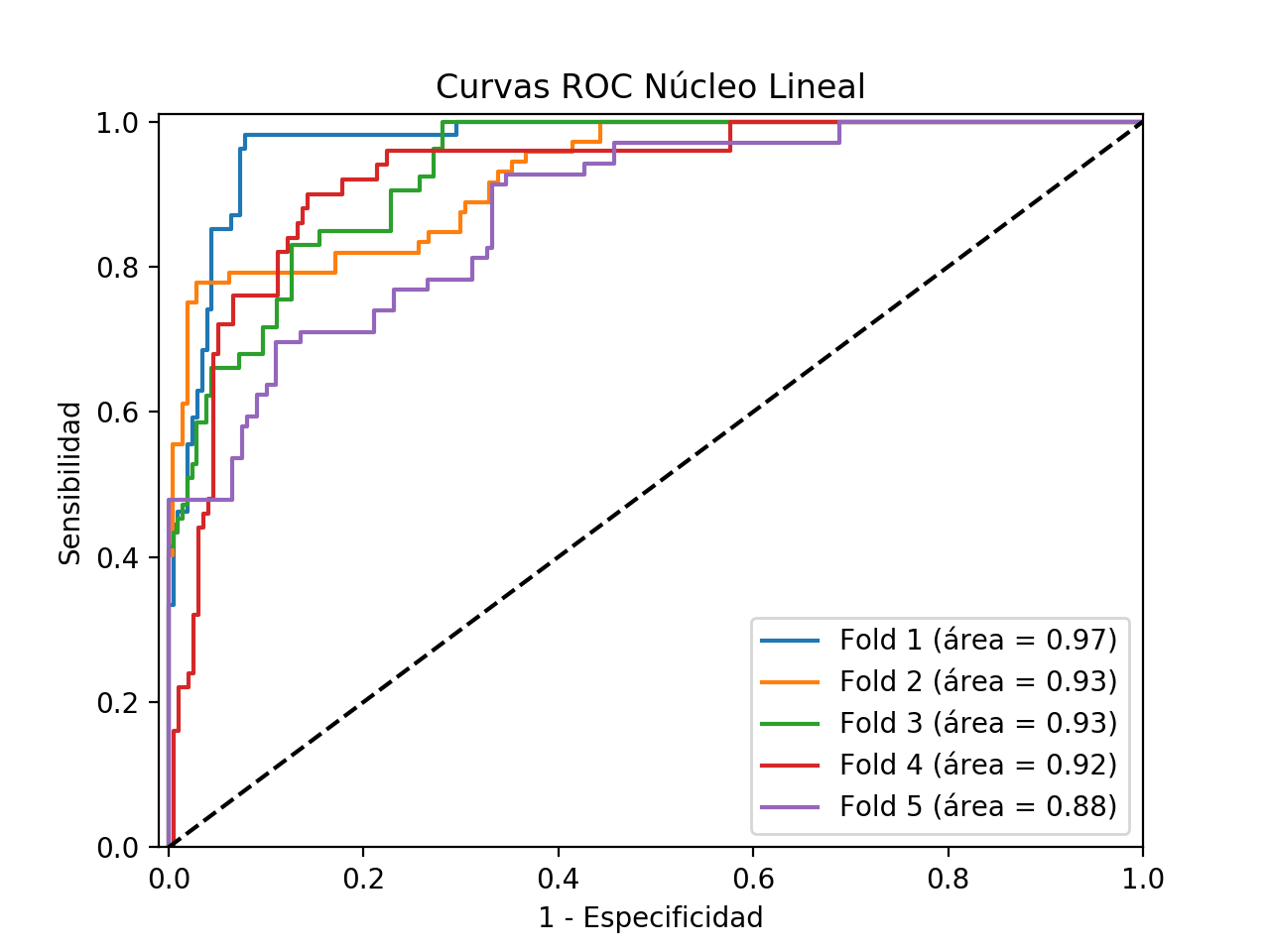
ROC curves
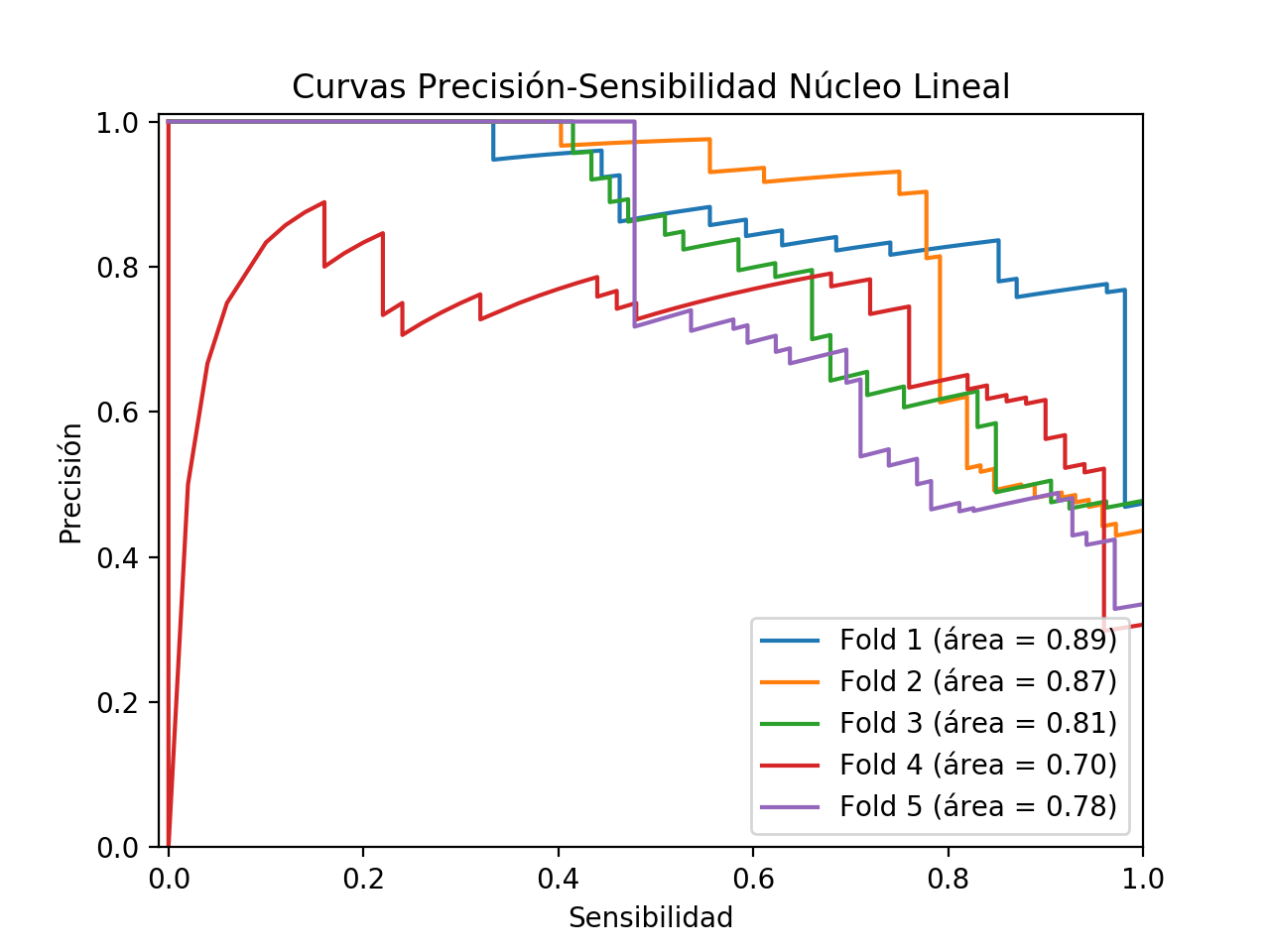
Precision/Recall curves
Confusion Matrices
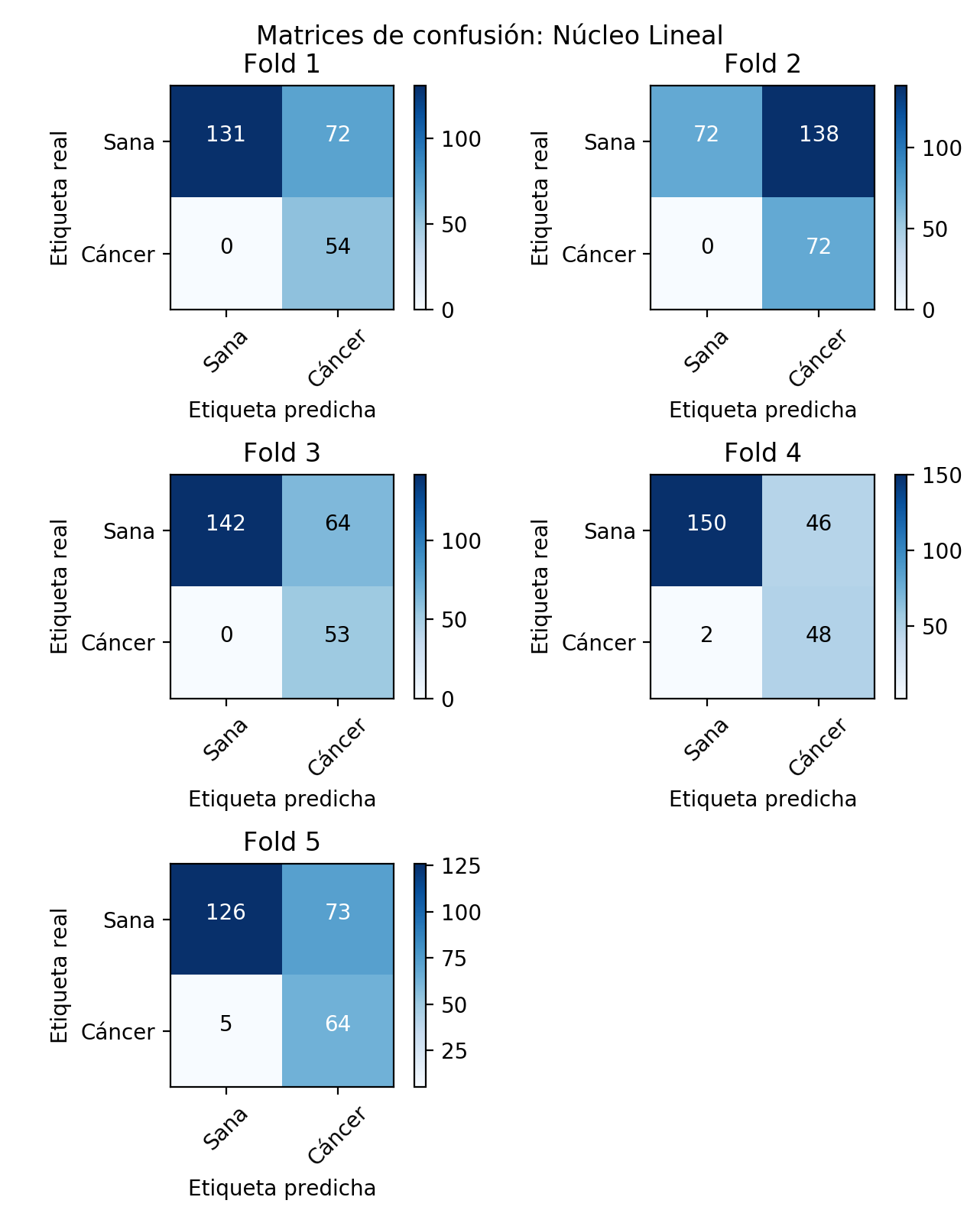
Similarly, the confusion matrices corresponding to each of the models generated in the cross validation are set out below.
Metrics by fold
Similarly, concrete metrics calculated from the confusion matrix are presented:
Commentary on the results for linear kernel
After having obtained each one of the graphs and metrics previously exposed, it is possible to say that in general terms, the Linear kernel has a somewhat irregular behavior. In the first place, it must be said that the goodness of the classification is not always good, as observed in the previous results. Thus, as can be seen in fold 2, for example, the accuracy of the classifier is around 50%, with a specificity close to 35%, but nevertheless with a sensitivity of 100%. Thus, in this case the classifier seems to be robust to false negatives, penalizing to a large extent the false positives, which in the case of a problem as critical as cancer detection, would prove to be a good feature. However, in this particular fold, it seems as if the model is not able to classify the negative class. The rest of the folds maintain a similar behaviour with respect to sensitivity, although in 4 and 5 certain false negatives are already sighted. However, it is in the predictive capacity of the negative class where the rest of folds have a much better performance than the 2.
Kernel Gaussian
As with the linear kernel, a series of metrics and graphs have been obtained for the Gaussian Gaussian kernel process results, which are presented below.
ROC curve and precision-sensitivity curve
It presents both the ROC curves obtained for each of the models generated in the different divisions of the data in the cross validation, as well as the precision and sensitivity curves.
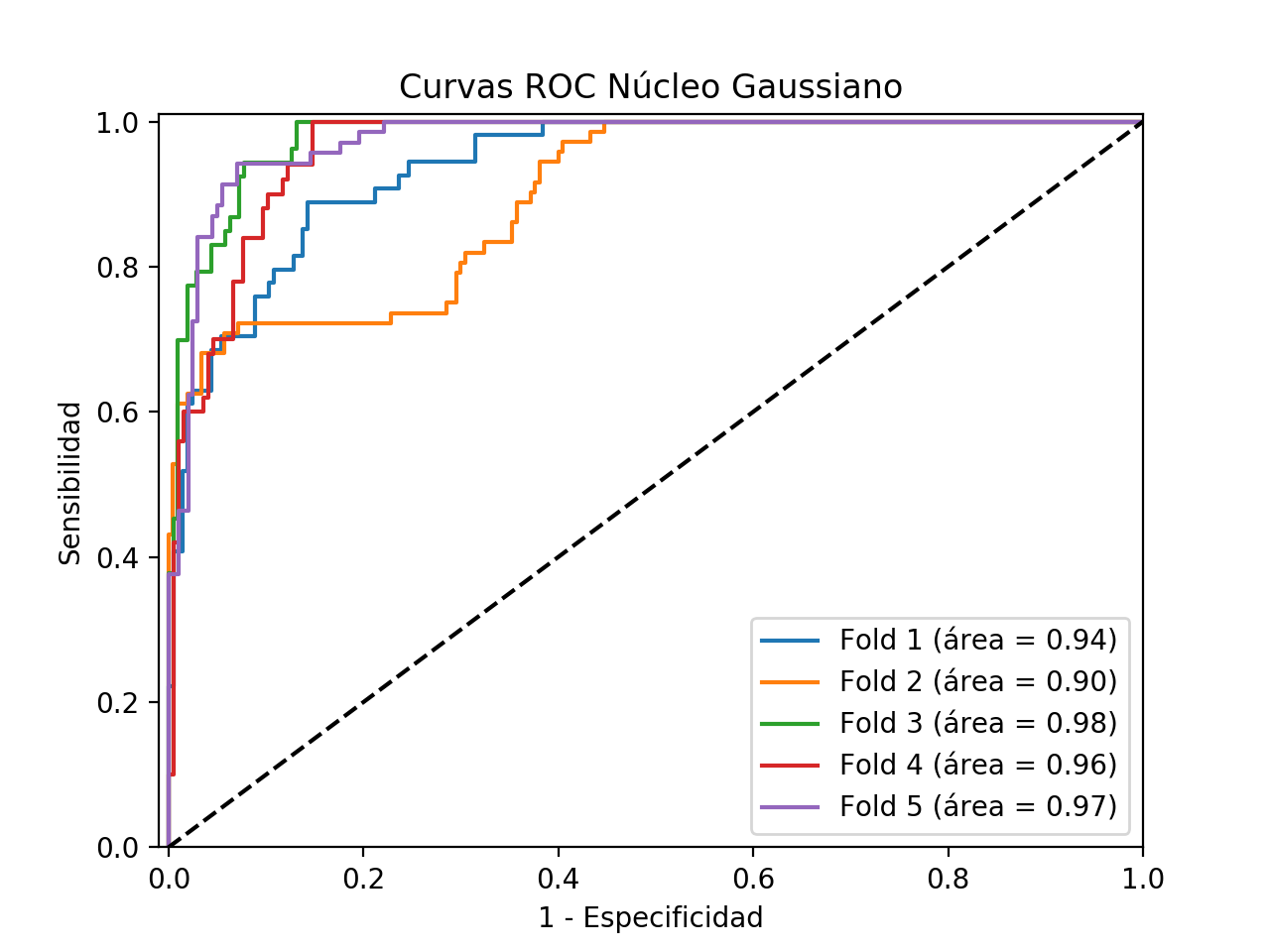
ROC curves
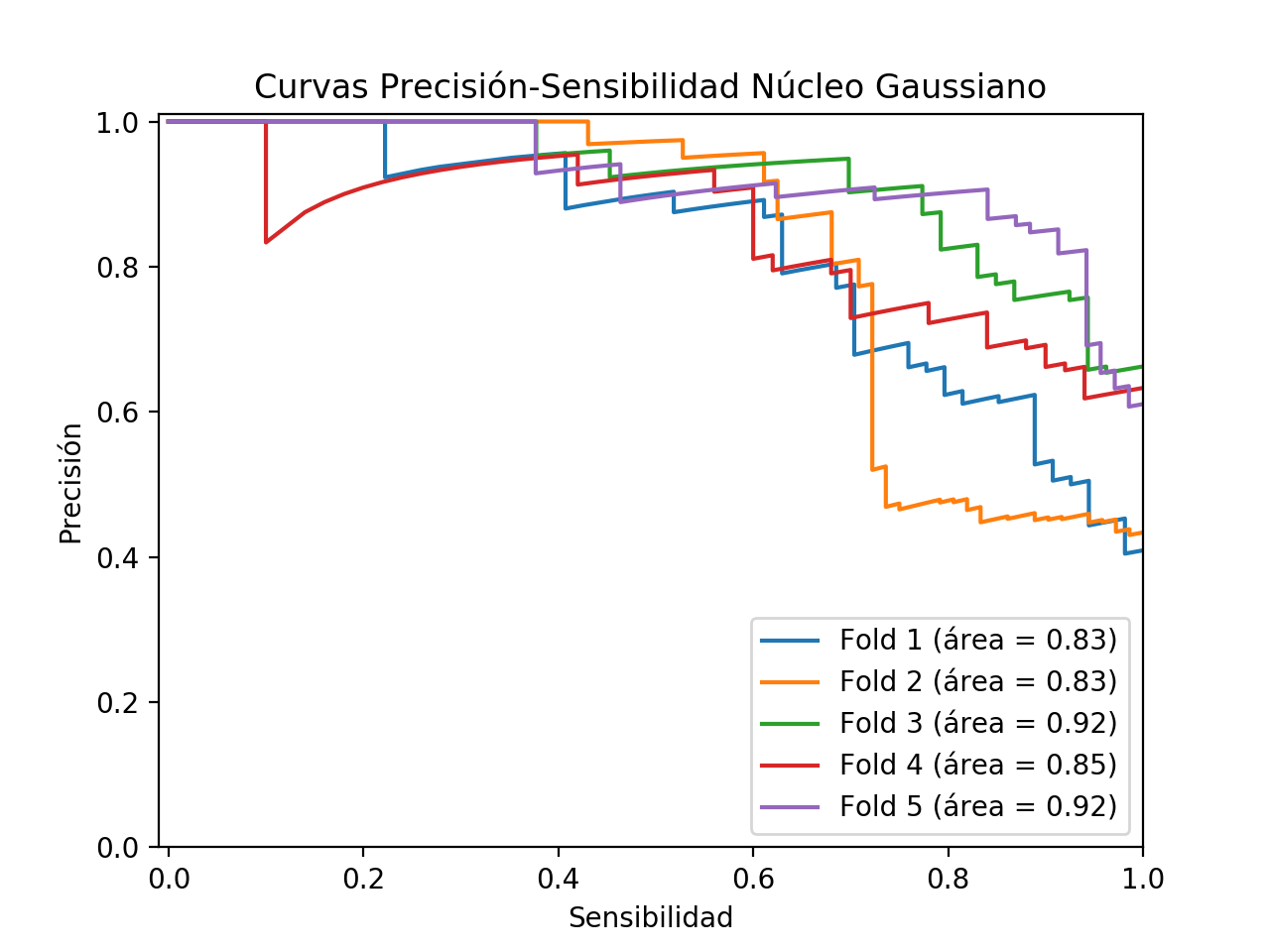
Precision/Recall curves
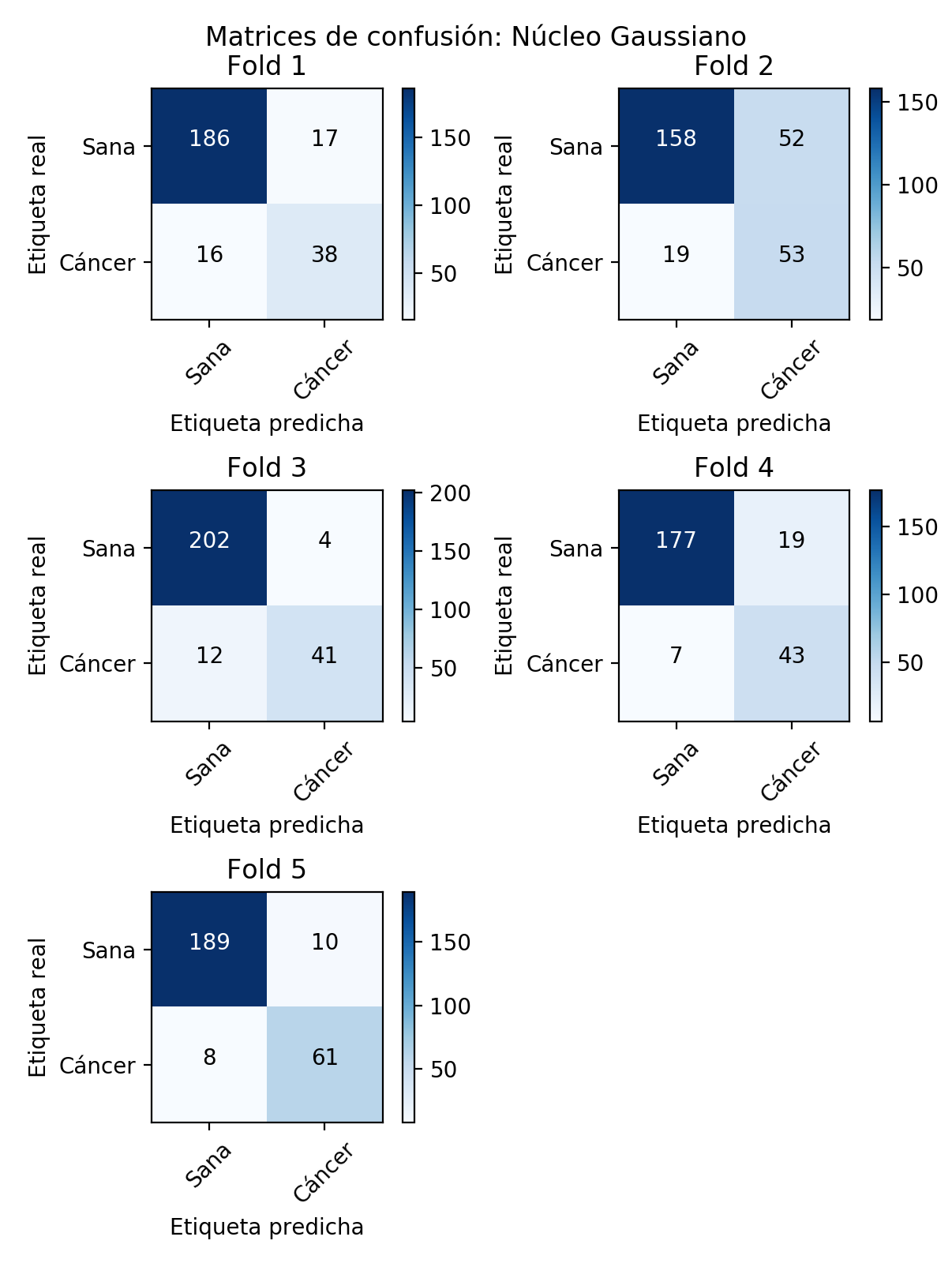
Confusion Matrices
Similarly, the confusion matrices corresponding to each of the models generated in the cross validation are set out below.
Metrics by fold
Similarly, concrete metrics calculated from the confusion matrix are presented:
Commentary on the results for Gaussian kernel
In general terms, these models with Gaussian kernel classify better than those obtained with linear kernel since their accuracy (Accuracy) so shows. However, as this is a highly unbalanced problem, we have to consider other measures that let us glimpse the extent to which our classifier is useful for solving our problem. Thus, we can observe how the classification of the negative class is generally good for all the models generated in the different folds, being the specificity of the models much better than those obtained in the models with linear kernel. Another fundamental aspect in a problem of these characteristics is the sensitivity of our models since as it can be appreciated, in these models with Gaussian kernel, this decreases considerably with respect to the results obtained with the linear kernel. This is a significant and very negative aspect, because on subjects that are sick, our classifiers would give them as healthy, having in this case a very high cost the erroneous classification. Thus, although both the specificity and precision of these models are better than those obtained with a linear kernel, the low sensitivity could make us consider the option of even discarding these models.